1. Introduction
The harmful substance Aflatoxin B1 (AFB-1), which has carcinogenic properties and could pose a major health crisis, has been identified as one of the greatest health risks that humans have faced throughout history (Qureshi et al., 2015). They are synthesized as secondary metabolites of toxigenic Aspergillus flavus, Aspergillus parasiticus, and Aspergillus nomius strains, and aflatoxin contamination may occur in various food commodities (Peltonen et al., 2001). If dairy products are contaminated with aflatoxin, the fermentation process may be disrupted, and unwanted textures and odor compounds may be produced in the product.
Aflatoxin exposure affects around 4.5 billion people, most of whom live in developing countries, and is largely unregulated. Mycotoxins can result in both acute and chronic intoxication depending on various parameters, such as intake levels, duration of exposure, toxin type, methods of action, metabolism, and defense mechanisms. Mycotoxins can be dangerous in four different ways: acute, long-term, mutagenic, and teratogenic (Kabak and Dobson, 2009). Acute mycotoxin poisoning occurs most frequently, having an immediate consequence leading to serious sickness that develops soon after consumption of food products containing the mycotoxins. Common side effects are deterioration of liver or kidney function, which in severe cases might result in death. (Dai et al., 2022; Pickova et al., 2021). Additionally, AFB-1 and its metabolites cause a variety of severe side effects, including toxicity to the liver, heart, spleen, brain, gut, skin, and testicles, as well as mutagenicity, teratogenicity, and carcinogenicity (Kabak and Dobson, 2009; Udomkun et al., 2017).
To control aflatoxin contamination, biological control involves the competitive exclusion of toxigenic strains by non-toxigenic strains (Udomkun et al., 2017). Recent research has shown that the utilization of probiotic lactic acid bacteria (LAB) strains (Enterococcus spp., Lactococcus spp., and Lactobacillus spp.) can effectively remove mycotoxins like aflatoxins, trichothecenes, and fumonisins from different food items before harvesting, processing, and storage (Deepthi et al., 2016). Several studies suggest that LAB can eliminate mycotoxins through enzymatic and adsorption mechanisms. The adsorption mechanism of LAB depends on the carbohydrate and protein properties of its cell wall, such as teichoic acid, where AFB1 binds through non-covalent interactions, including Van der Waals forces, hydrogen bonds, and electrostatic interactions. The integrity of the LAB cell wall is crucial for AFB1 adsorption, as the number of binding sites in the cell wall is essential for its binding capacity for both viable and nonviable (heat-treated) organisms (Afshnar et al., 2020; Emadi et al., 2021; Pflieger et al., 2015). Enzymatic treatment involves two enzymes, 17-hydroxy-steroid dehydrogenase, and carboxypeptidase A, which are believed to degrade mycotoxins. These enzymes, produced by LAB, play a key role in detoxification by cleaving the furofuran and lactone rings, structures essential for the toxigenicity of aflatoxins, leading to the production of aflatoxicol, a metabolite of AFB1 (Afshnar et al., 2020; Emadi et al., 2021). In other studies, LAB was also described to have a potential adsorbent capability for AFB-1 observed in both in vitro and in vivo methods (Zolfaghari et al., 2020). In a previous study, Lactobacillus plantarum 1, a lactic acid bacteria strain, was isolated from coconut toddy, a drink common in the Philippines that is usually called “tuba” (Hinay et al., 2022). In this study, we molecularly characterized the isolated lactic acid bacteria and evaluated their binding capacity against AFB-1. The isolated Limosilactobacillus fermentum provides a promising AFB-1 binding capacity and requires further study to develop its possible application in the food industry.
2. Materials and methods
The fermented coconut toddy (approximately 60 days old) samples were procured from Hijo, Maco, Davao de Oro, Philippines, and stored in a sterile container to ensure that there was no contamination by other microorganisms. The sample was placed inside a transportation box filled with ice to ensure the viability of the lactic acid bacteria.
The isolation of LAB strains was obtained from three different fermented coconut toddy samples. The fermented coconut toddy (10 mL) was mixed with 90 mL of sterile saline solution (0.50% NSS) and serially diluted to 106. A 0.1 mL diluted sample was spread on De Man Ragosa and Sharpe (MRS) agar (Hi-Media Laboratories, Thane, India) and incubated for 72 h anaerobically at 37°C. Successive dilution provides cells at a sufficiently low density such that single cells are physically isolated spatially to produce recognizable individual colonies. Individual colonies were subjected to molecular identification.
Isolated LABs were grown at 37°C under anaerobic conditions in De Man, Rogosa, and Sharpe (MRS) broth (Hi-Media Laboratories, Thane, India) for 72 h. Total genomic DNA was extracted from cell pellets (centrifugation at 8,000 ×g for 5 min) using the innuPREP DNA Mini Kit (Innuscreen GmbH., Germany) according to the manufacturer’s instructions. Quantity and purity of DNA were spectrophotometrically checked using a NanoDrop ND-1000 (Thermo Scientific USA). Extracted DNA samples were stored at 20°C for subsequent application.
Complete 16S rRNA genes were PCR-amplified with primer pairs 27 F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492 R (5’-GGTTACCTTGTTACGACTT-3’) (Heilig et al., 2002). A 25 μL PCR mixture contains 0.5 μL Taq DNA polymerase, 400 M MgCl2, 0.5 μL of each primer, 4 μL of dATP, dGTP, dCTP, dTTP, 2.5 μL of reaction buffer, and 4 μL of DNA template. Thermal cycling conditions were 94°C for 2 min; 32 cycles of 45 s denaturation at 94°C, 45 s annealing at 55°C, and 1.25 min extension at 72°C; final extension at 72°C for 5 min.
All amplified PCR products were purified using the Macherey-Nagel Purification Kit (Macherey-Nagel, Germany) according to the manufacturer’s instructions and were electrophoresed on an agarose gel and visualized using an ultraviolet transilluminator (GelStudio - Analytik Jena GmbH., Jena, Germany) (Park et al., 2005).
Purified PCR products were sequenced using a SeqStudio Genetic Analyzer Applied Biosystem (Thermo Fisher Scientific, USA). The obtained nucleotide sequences of 16S rRNA were searched using BLAST in GenBank (National Center for Biotechnology Information, Rockville Pike, Bethesda, USA) using the advanced BLAST similarity search option. Nucleotide sequences were aligned and compared with other nucleotide sequences retrieved from GenBank using Clustal W, and the phylogenetic tree was generated using the neighbor-joining method in MEGA software (version 6.0; Biodesign Institute, Tempe, USA) (Kumar et al., 2018).
The identified LAB isolate was initially evaluated for possible AFB-1-binding capacity using phylogenetic analysis and compared with known LAB with AFB-1-binding capacity (Haskard et al., 2001). Phylogenetic analysis was performed following the protocol mentioned above.
The isolated LABs were grown in MRS broth (Hi-Media Laboratories., Thane, India) for 72 h under anaerobic conditions. The bacterial concentration was standardized at an optical density (OD) of 1.0 (600 nm) using a UV-VIS spectrophotometer to obtain an approximate density of 109 cells. After centrifugation at 3,500 rpm for 15 min at room temperature (25°C), the pellet containing live cells was resuspended in the same volume of 1× PBS buffer (pH 7.4). Subsequently, 200 μL of PBS was added to the pellet to form a suspension. LAB suspensions were separated into live and heat-treated bacterial cells. Briefly, 200 μL of the bacterial sample was suspended in a tube. Heat-treated bacterial cells were then prepared by boiling the suspension, which contained live cells, at 100°C for 1 h in a water bath (Haskard et al., 2001).
The binding capacity of Limosilactobacillus fermentum to AFB-1 was analyzed using an enzyme-linked immunosorbent assay (ELISA) Kit (Shenzhen Lvshiyuam Biotechnology Co., Ltd., Shenzhen, China). Briefly, 50 μL of AFB-1 (1.64 ppb) was added to the cell suspension and incubated for 1 h at 37°C. The AFB-1-Limosilactobacillus suspension (100 μL) was then dispensed into each well, and 50 μL of the enzyme conjugate and antibody working solution were added, and the wells were incubated at 25°C for 30 min. The plates were washed four times using 250 μL washing buffer, and 50 μL of Substrate A and Substrate B were added and incubated at 25°C for 15 min. After incubation, 50 μL of stop solution was added, and the absorbance of the wells was read at 450 nm to determine the OD value to determine the binding capacity of L. fermentum (Liew et al., 2018).
3. Results and discussion
Incubation of the collected fermented coconut toddy for 72 h at 37°C anaerobically produced uniform phenotypic colony characteristics on the surface of MRS agar. Three MRS agar plates were screened for small, round, matte, and white colonies, of which three bacterial colonies were subjected to molecular characterization.
Fig. 1A confirmed the genomic DNA isolated from bacterial samples, in addition, Fig. 1B presents PCR fragments of ~700 bp in size that were amplified for all isolates using specific primers. Of the three isolates, amplified fragments were purified (Fig. 1C) for sequence analysis.
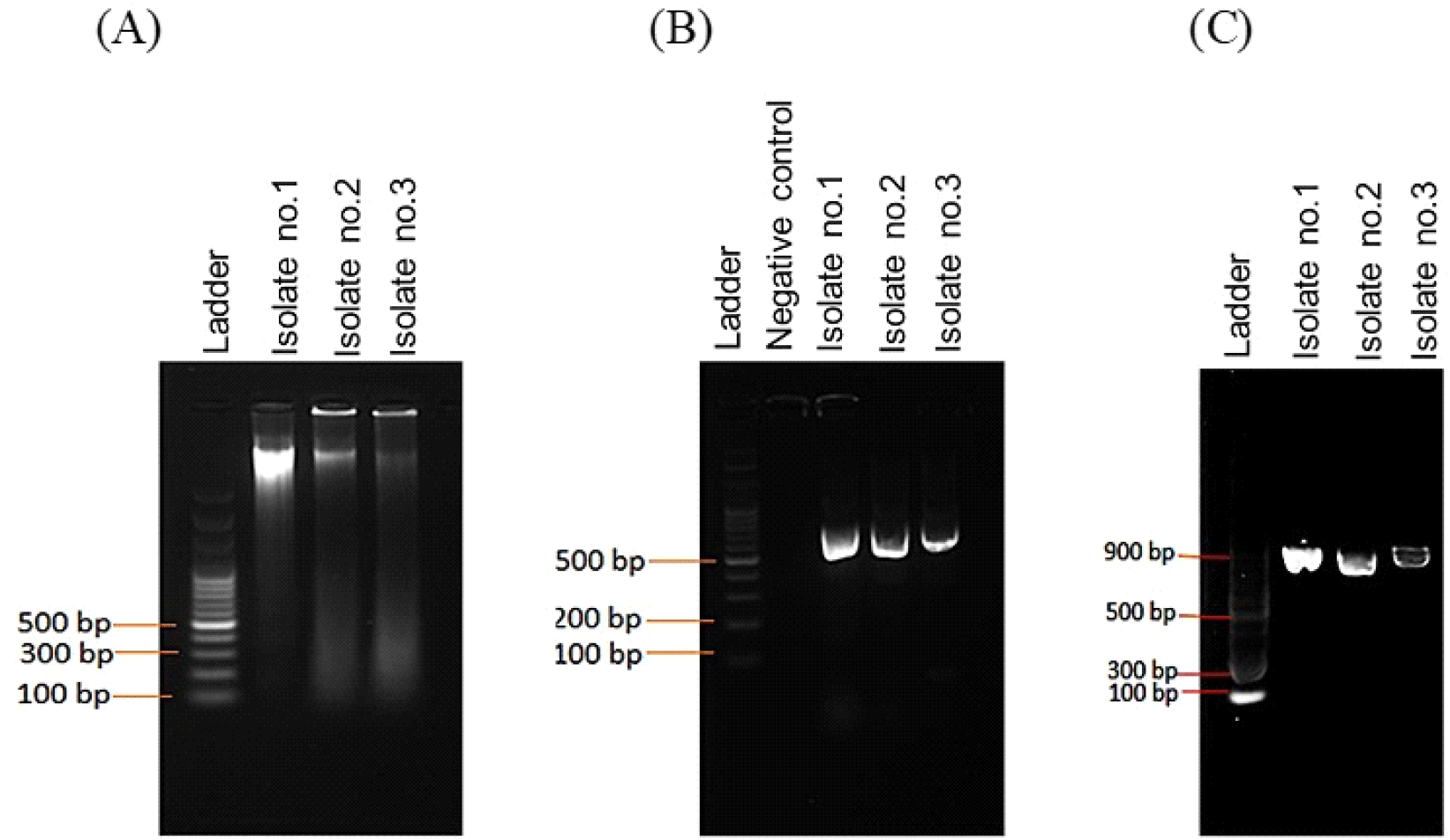
The three isolates were subjected to species identification by MEG7/NCVI-BLSTN. Isolates 1 and 2 have 100% similar consensus sequences, except for isolate 3. The isolate 1 16sRNA sequence, identified as Limosilactobacillus fermentum, was deposited in the International Nucleotide Sequence Database under the accession number OR536351.1. The 16 s rRNA sequences of the species used in the phylogenetic analysis were obtained from GenBank as of September 4, 2023. In Fig. 2, these sequences were aligned using MAFFT v.7.520 with default parameters (Katoh and Standley, 2013). Following multiple sequence alignment, poorly aligned regions were trimmed using TrimAI v.1.2 (Capella-Gutiérrez et al., 2009). A maximum likelihood (ML) phylogenetic tree of the genome was inferred using IQTree v.2.2.2.7 with a GTR substitution model and gamma-invariant sites, with 1000 bootstrap replicates. The sequence alignment was viewed using JalView v.2.11.2 and was edited using FigTree v.1.4.47 (Minh et al., 2020; Rambaut, 2010; Waterhouse et al., 2009).
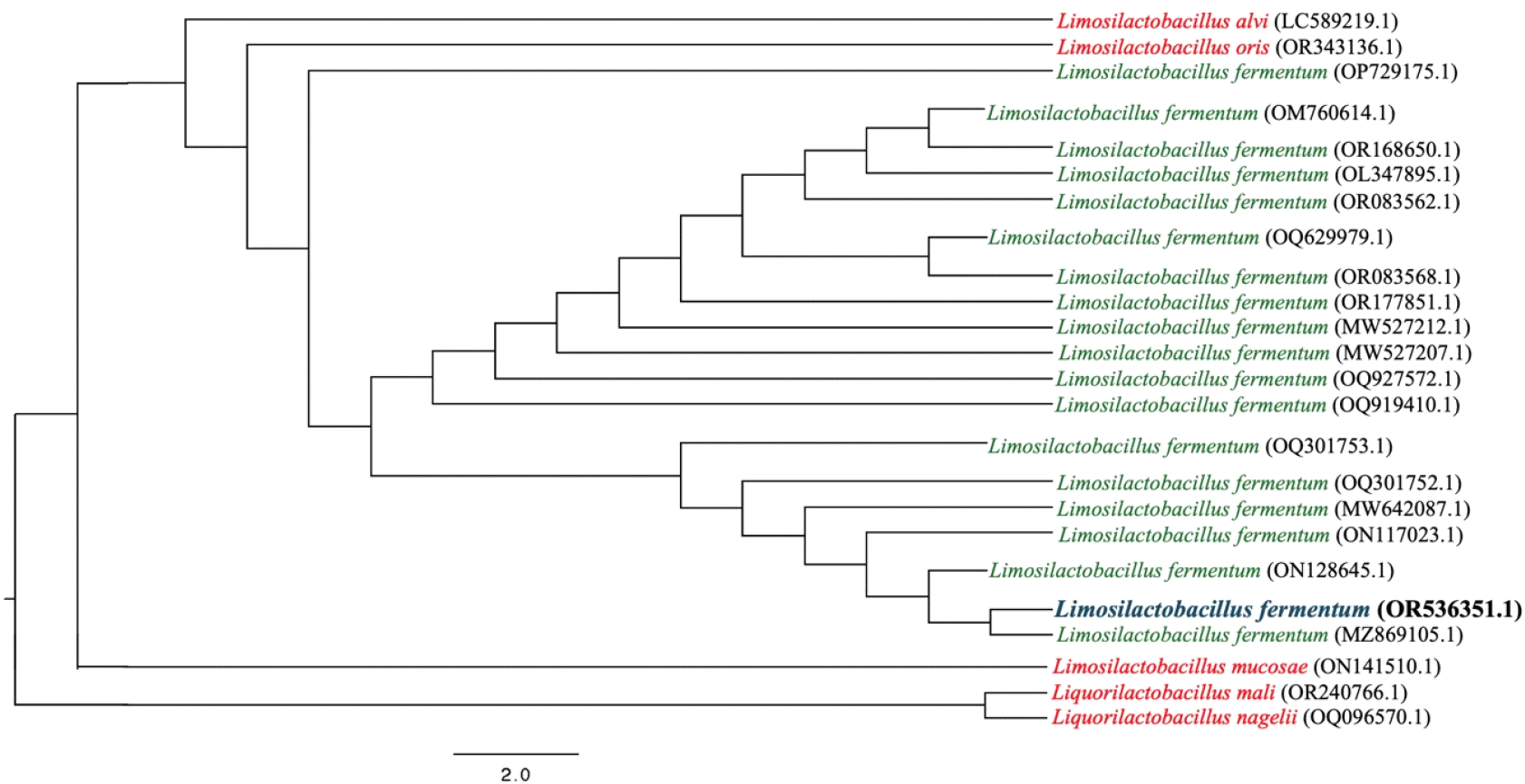
Fig. 3 shows the phylogenetic analysis that was conducted to compare the isolated Lactobacillus fermentum strain with other lactic acid bacteria known for their AFB-1 binding capacities. The analysis revealed that L. fermentum shares a close genetic relationship with Propionibacterium freudenreichii subsp. shermanii. This finding is particularly significant, given that P. freudenreichii subsp. shermanii has previously been reported to exhibit substantial AFB-1 binding activity, with binding efficiencies of 22.3% in live cells and 67.3% in heat-treated cells (Haskard et al., 2001). The close phylogenetic relationship observed suggests that genetic factors may underlie, at least in part, the capacity of these microorganisms to bind AFB-1. To experimentally validate this hypothesis, the AFB-1 binding capacity of the isolated L. fermentum strain was assessed using an ELISA-based assay.
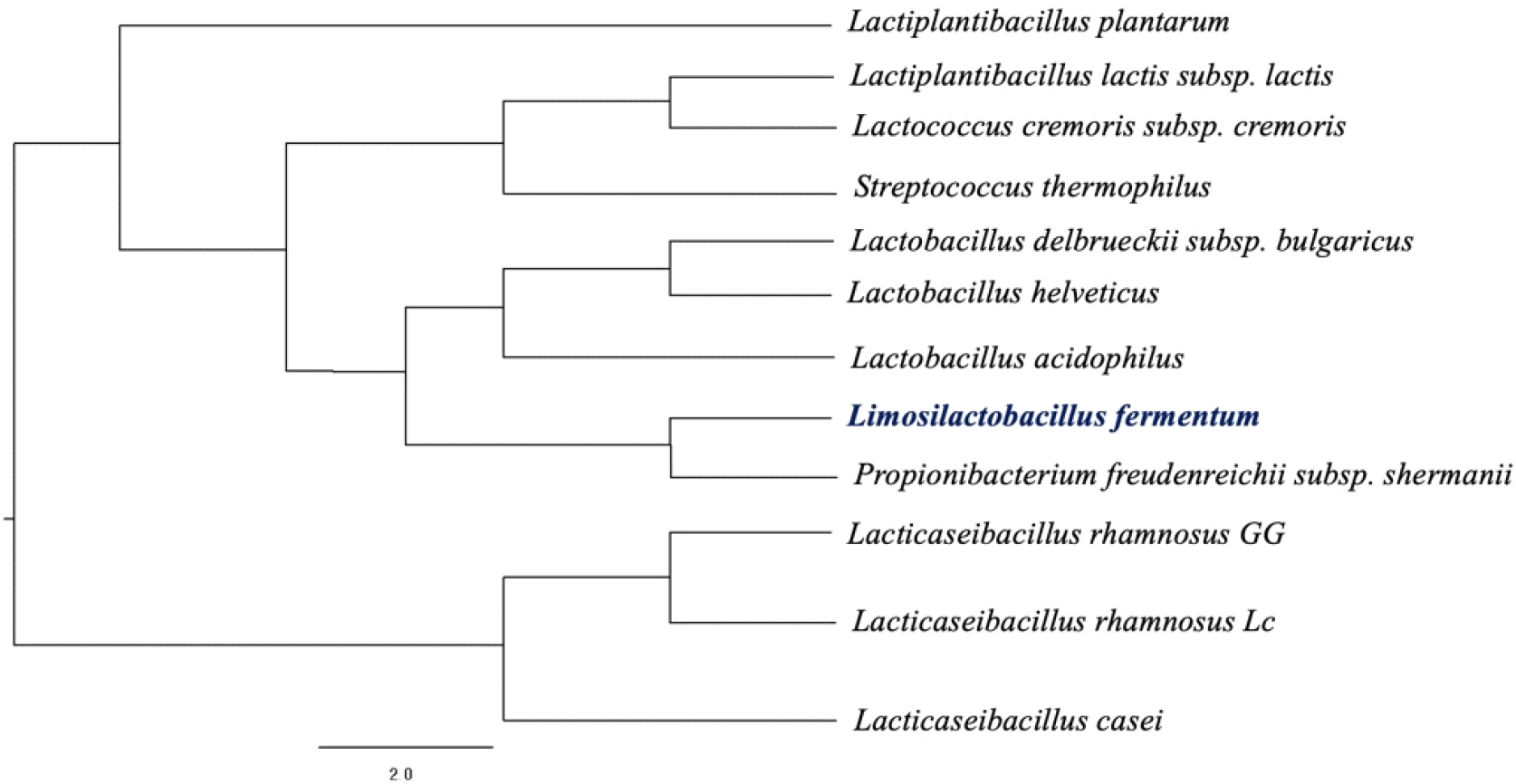
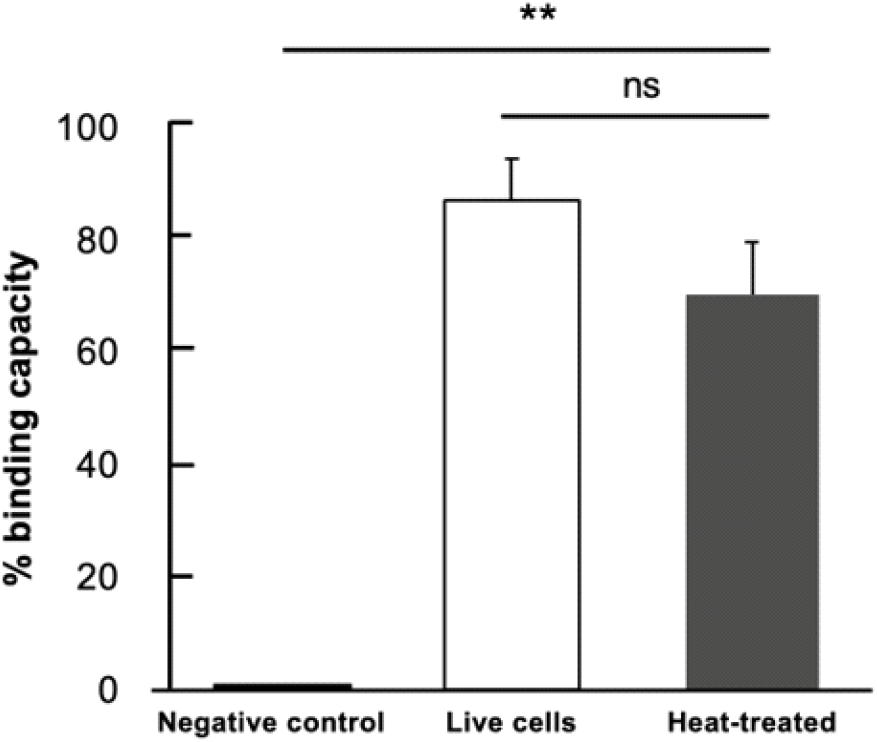
Fig. 4 displays the ELISA assay results for both live and heat-treated bacteria, revealing that most of the AFB-1 is bound to the bacterial surface, with a binding capacity of 87.30±7.29% for live cells. The heat-treated sample, however, shows a lower binding capacity of 70.49±9.59%, which is still comparable to the binding capacity of live bacteria (p= 0.2217), though significantly reduced. The large margin of error observed in the data can be explained by the sample’s heterogeneity and the use of non-purified samples. Specifically, live organisms possess intact cell walls and complete binding sites, which enhance the binding capacity of AFB-1. In contrast, heat treatment leads to structural alterations in the bacterial cell wall, decreasing the number of available binding sites and resulting in a lower binding capacity. This finding is consistent with previous studies (Afshnar et al., 2020; Emadi et al., 2021; Pflieger et al., 2015), which emphasize the importance of the bacterial cell wall’s structural integrity for binding AFB-1. Furthermore, the use of non-purified samples introduces additional variables, such as polar matrix components present in the culture media (Bata-Vidacs et al., 2020) and cellular debris that interfere with the binding of AFB-1 to the cell wall binding sites. Although these factors contribute to variability, they reflect real-world conditions where sample purification may not always be feasible. Despite the variability, the binding values (ranging from 70-87%) remain biologically meaningful, demonstrating the substantial interaction capacity of the sample. This suggests that L. fermentum maintains its binding capability even after being subjected to heat treatment, although with a slight reduction compared to that of live cells. Moreover, this binding capacity was higher than that of 12 lactic acid bacteria evaluated in another study (Haskard et al., 2001). Remarkably, live cells most efficiently removed the highest amount of AFB-1 from the solution. This suggests that the surface components of L. fermentum are involved in binding to AFB-1. The treatment of bacteria with heat may affect the AFB-1 binding mechanism; however, heat-treated L. fermentum also provided the ability to bind AFB-1, similar to live bacteria. The slight difference in the binding capacity of AFB-1 between the two preparations could be due to cell wall polysaccharides and peptidoglycan, which are the two main elements responsible for the binding of mutagens to LAB. Both components are expected to be significantly affected by the heat treatment (Hatakeyama et al., 2011; Zhang and Ohta, 1993).
4. Conclusions
In conclusion, the fermented coconut toddy analyzed in the current study was found to be a potential source of lactic acid bacteria (LAB), particularly Limosilactobacillus fermentum. This identification was established through 16S rRNA gene sequencing and phylogenetic analysis. These techniques are valuable tools for the precise identification and molecular characterization of lactic acid bacteria. In addition, phylogenetic prediction analysis can serve as a basis for experimental validation, particularly in assessing the AFB-1 binding capacity of the identified LAB strain. This finding suggests a potential application of L. fermentum in mitigating aflatoxin contamination, highlighting its significance in food safety and quality control efforts.










