Introduction
Salmonella are gram-negative, facultative anaerobic, and mesophilic foodborne pathogen (Allerberger et al., 2003; FDA, 2012). Generally, Salmonella can grow and survive in animal-originated foods and fresh products (Cetin et al., 2020; Eng et al., 2015; Hyeon et al., 2011; Kim et al., 2018). In 2015, Salmonella was the second-most common cause (34%) of foodborne outbreaks in the United States, after norovirus (37%). Pork, seeded vegetables, and vegetable row crops were commonly responsible for these outbreaks (CDC, 2017).
Kimchi is a traditional side dish and the most widely consumed fermented food in South Korea, served by most restaurants and food services (Rhee et al., 2011). Kimchi has various health benefits conferred by the presence of lactic acid bacteria (LAB), organic acids, and bacteriocins (Park et al., 2014). LAB in kimchi are known to have antimicrobial effects against Salmonella, Staphylococcus aureus, Bacillus cereus, and Escherichia coli (Ahn et al., 2003). Although, kimchi is considered microbiologically safe, the safety and microbial risk of kimchi can vary, depending on several factors such as the fermentation period or the components of the raw materials used. Lee et al. (2017) showed that fresh kimchi has a low concentration of LAB, which might be insufficient to suppress pathogens. In addition, kimchi can be cross-contaminated by Salmonella and other foodborne pathogens during manufacturing when the napa cabbage is soaked in the NaCl solution (Yu et al., 2012). Thus, the causes of unexpected kimchi contamination outbreaks remain unclear (e.g., contamination of the raw ingredients of kimchi or contamination during manufacturing) (Cho et al., 2014; Patra et al., 2016).
To investigate the kinetic behavior of foodborne pathogens, predictive models have been used (Ha et al., 2016; Ha et al., 2019; Kim et al., 2017; Lee et al., 2014; Lee et al., 2019; Yoon et al., 2006). In these models, changes in bacterial cell counts depend on changes in time and kinetic parameters such as lag phase duration (LPD) and growth rate, which are calculated using a primary model. Secondary models are used to describe the effects of environmental factors, such as temperature, pH, and water activity, on kinetic parameters (Lee et al., 2014; Lee et al., 2015).
The objective of this study was to develop predictive models to describe the kinetic behavior of Salmonella in napa cabbage kimchi during fermentation.
Materials and methods
Raw ingredients, including napa cabbage (heading type), Korean white radish (Joseonmu), chives, ground garlic, ground ginger, and Korean red pepper powder were purchased from a grocery store in Seoul, South Korea. All ingredients were produced in Korea. The napa cabbage was salted with 10% NaCl solution for 3 h and drained. The ingredients were first cut; 4-cm-long pieces of napa cabbage leaves, 5-cm-long pieces of white radish, and 5-cm-long pieces of chives were prepared. The ingredients (1 kg of salted napa cabbage with 172 g sliced white radish, 16 g chives, 3.2 g ground ginger, 24 g ground garlic, 22 g powdered red pepper, and 2 g sugar) were then mixed thoroughly (KTKP, 2018; RDA, 2008). One-kilogram portions of napa cabbage kimchi were placed in a zip lock bag (30×45 cm).
Five Salmonella strains (S.enterica serovar Typhimurium NCCP10812, S.enterica serovar Agona NCCP12231, S.enterica serovar Enteritidis NCCP12243, S.enterica serovar Montevideo NCCP10141, and S. enterica KACC11595) were used in this study. Single colonies of Salmonella were cultured in 10 mL tryptic soy broth (TSB) (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) at 37°C for 24 h. One hundred microliter aliquots of the cultures were transferred into 10 mL of fresh TSB and sub-cultured under the same conditions. The sub-cultured Salmonella strains were mixed, and 50 mL of the Salmonella mixture was centrifuged at 1,912 ×g and 4°C for 15 min. The pellet was washed twice with phosphate-buffered saline (PBS) (pH 7.4; 0.2 g of KH2PO4, 1.5 g of Na2HPO4·7H2O, 8.0 g of NaCl, and 0.2 g of KCl in 1 L of distilled water) and resuspended in PBS to obtain 6-7 log CFU/mL of inoculum. Twenty milliliters of the sub-cultures were inoculated onto kimchi from each zip lock bag to obtain 4-5 log CFU/g. Zip lock bags were massaged 30 times and placed into airtight plastic containers. The airtight plastic containers were stored at 4°C, 10°C, 15°C, 25°C, and 30°C for 48-384 h for fermentation.
Fifty grams of kimchi samples from each ziplock bag (n=4) were aseptically removed with a flame-sterilized tong, and placed in filter bags (3M, St. Paul, MN, USA) containing 100 mL of 0.1% buffered peptone water (BPW) and pummeled (BagMixer; Interscience, St. Nom, France) for 1 min. The homogenates were serially diluted with 0.1% BPW, and the diluents were plated on xylose lysine deoxycholate (XLD) agar (Becton, Dickinson and Company) and Lactobacilli MRS agar (Becton, Dickinson and Company) for the isolation of Salmonella and lactic acid bacteria, respectively. All plates were incubated at 37°C for 24-48 h. The pH of the kimchi samples was measured using a pH meter (Thermo Fisher Scientific, Waltham, MA, USA).
The Baranyi model (primary model) was fitted to Salmonella cell counts to calculate kinetic parameters using the DMfit program (Institute of Food Research, Norwich, UK) to fit the growth curve (Baranyi and Roberts, 1994). The kinetic parameters were described using primary models to calculate the LPD and maximum specific growth rate (μmax; log CFU/g/h) in the growth phase and death rate (DR; log CFU/g/h) in the death phase. The relationship between kinetic parameters and fermentation temperature was analyzed using a polynomial (secondary) model developed using SigmaPlot 10.0 (Systat Software, San Jose, CA, USA) based on the following equation:
where Temp is the fermentation temperature, and a, b, and c are constant values. To simulate the growth of Salmonella under varying temperatures, a dynamic model was developed in accordance with the developed primary and secondary models using the equation developed by Baranyi and Roberts (1994). The kimchi samples inoculated with Salmonella were stored at various temperatures (4°C, 15°C, and 25°C), and the temperature was changed twice every 24 h. The actual temperatures were measured using a data logger (Testo 174H, Testo, Lenzkirch, Germany).
To evaluate the performance of the developed model, Salmonella-inoculated kimchi samples were stored at 20°C and 27°C, and the cell counts of Salmonella were obtained as described above. These cell counts were used as the observed values which were then compared with the predicted values obtained through the simulation from the developed models at 20°C and 27°C. The differences between the observed and predicted values were obtained by calculating the root mean square error (RMSE) using the following equation:
Similarly, to validate the simulation of Salmonella cell counts predicted by the dynamic model under varying temperatures, Salmonella-inoculated samples were stored at 4°C, 15°C, and 25°C, and the temperature settings were changed twice every 24 h. The temperature changes were recorded using a data logger (Testo 174H, Testo). The recorded temperatures were further used for the simulations in the dynamic model. During storage, Salmonella cell counts were enumerated as described above. The cell counts were then compared to the data predicted by the dynamic model simulation.
Kinetic parameters at each temperature were analyzed using the PROC GLM procedure of SAS® v.9.3 (SAS Institute, Cary, NC, USA). Least square (LS) means for each kinetic parameter were compared using pairwise t-tests at α=0.05.
Proliferation of Salmonella in kimchi decreased as the population of LAB increased and as pH decreased, regardless of temperature (Fig. 1). However, Salmonella cell counts in kimchi were constant at 4-15°C, but increased in the early phase of fermentation at 25°C and 30°C, and subsequently decreased thereafter (Fig. 1D-E). Once napa cabbage kimchi is prepared, it is kept at room temperature for 12-24 h to accelerate fermentation. It is then either consumed or stored at 4°C. According to the Salmonella cell count data, consumption or storage at 4°C after 12-24 h of fermentation at room temperature may increase the risk of Salmonella foodborne illness because Salmonella cell counts increased during the 12-24 h fermentation period, and remained constant during storage at 4°C.
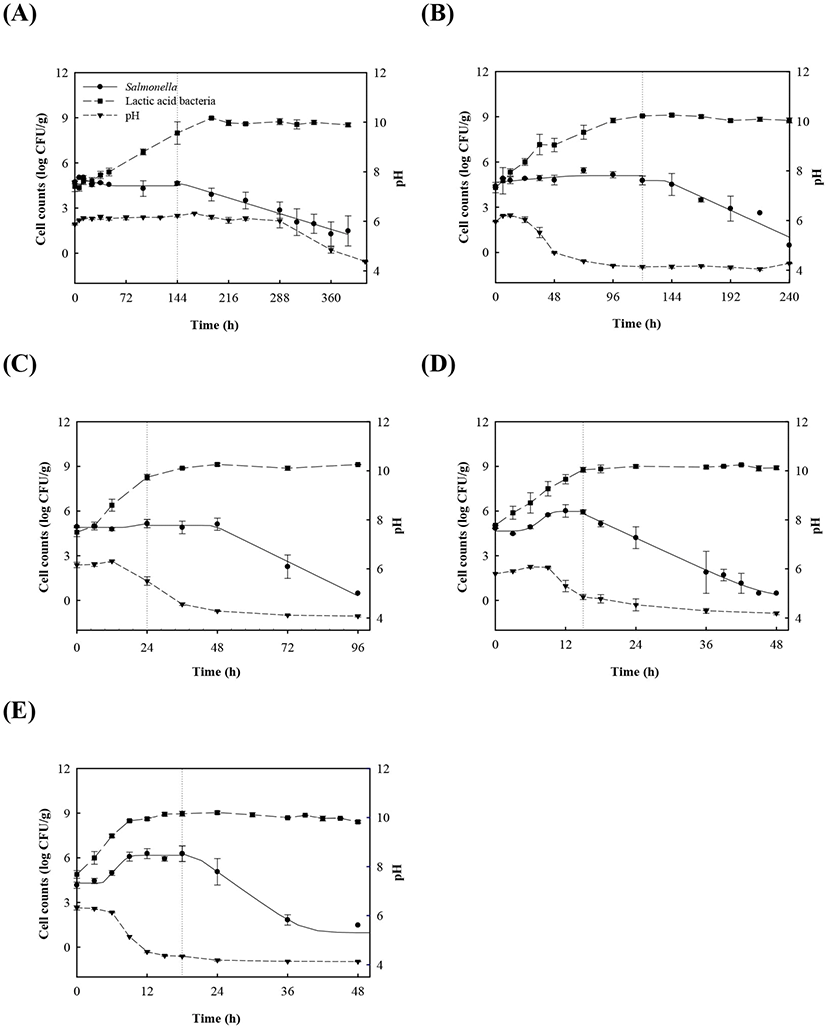
LAB counts increased from 4.7±0.6 to 8.7±0.3 log CFU/g, and pH decreased from 6.0±0.2 to 4.2±0.1 during fermentation. As the temperature increased, the LAB growth rate increased, and the pH decreased rapidly (Fig. 1), as described above. When increased to approximately 9 log CFU/g in kimchi, LAB showed strong antimicrobial activity against Salmonella by decreasing the pH and producing antimicrobials. Salmonella death might be caused by antimicrobial substances, lactic acid, or because of low pH generated by LAB during fermentation (De Keersmaecker et al., 2006; Park et al., 2005). During kimchi fermentation, the cell counts for Salmonella remained either constant or increased and subsequently decreased (Fig. 1). Thus, primary models were developed separately for the growth and death phases (Table 1 and Fig. 1). In the growth phase, LPD was 180.00, 120.00, 16.42, 5.85, and 4.81 h at 4°C, 10°C, 15°C, 25°C, and 30°C, respectively (Table 1). However, μmax [0.000 (4°C), 0.007(10°C), 0.033 (15°C), 0.302 (25°C), and 0.592 (30°C) log CFU/g/h] showed increment with temperature (p<0.05) (Table 1). The Salmonella cell counts then started to decrease after 180 h at 4°C, 120 h at 10°C, 24 h at 15°C, and 17 h at 25°C and 30°C after fermentation (Table 1). In the death phase, SP decreased [14.95 (4°C), 19.02 (10°C), 22.84 (15°C), 0.00 (25°C), and 1.04 h (30°C)], and DR increased [−0.018 (4°C), −0.037 (10°C), −0.096 (15°C), −0.149 (25°C), and −0.306 (30°C) log CFU/g/h] (p<0.05) as the temperature increased. The lower fermentation temperatures showed longer LPD, lower μmax, and lower DR. Secondary models were developed for the kinetic parameters (LPD, μmax, ITD, SP, and DR) with a polynomial model (R2=0.606−0.956) (Fig. 2). Validation of the performance of the developed predictive models was confirmed through an additional experiment conducted at 20°C and 27°C, where the RMSE values were 0.350 (growth phase) and 0.428 (death phase). This result indicates that the developed models were appropriate for predicting the kinetic behavior of Salmonella in kimchi. In addition, a dynamic model was developed to demonstrate the changes in Salmonella cell counts in kimchi at varying temperatures (4-25°C). In simulations with the dynamic model, Salmonella cell counts increased from 4.7 log CFU/g to 6.9 log CFU/g after 46 h, and then decreased to 0.8 log CFU/g after 135 h of fermentation. This dynamic model also showed that Salmonella can grow after the initiation of fermentation, as shown in the primary models (Fig. 3).
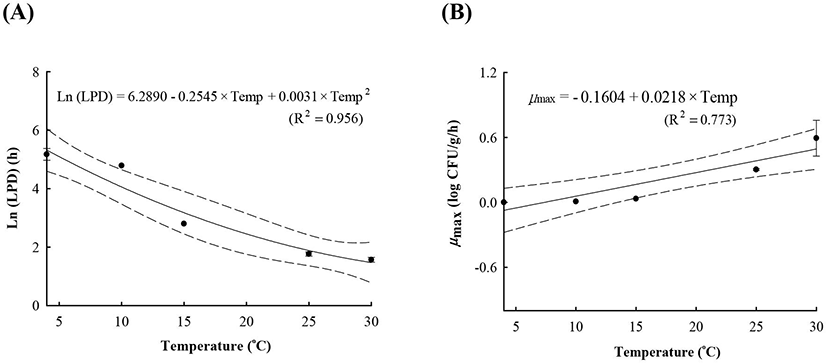
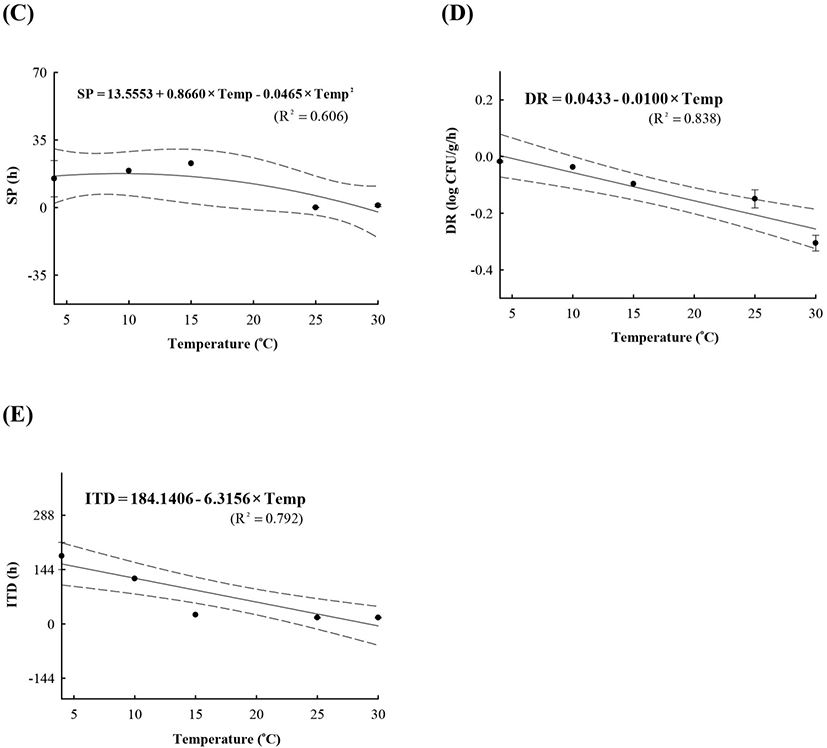
A, lag phase duration (LPD); B, maximum specific growth rate (μmax); C, shoulder period (SP); D, death rate (DR); E, initiation time of decrease (ITD).
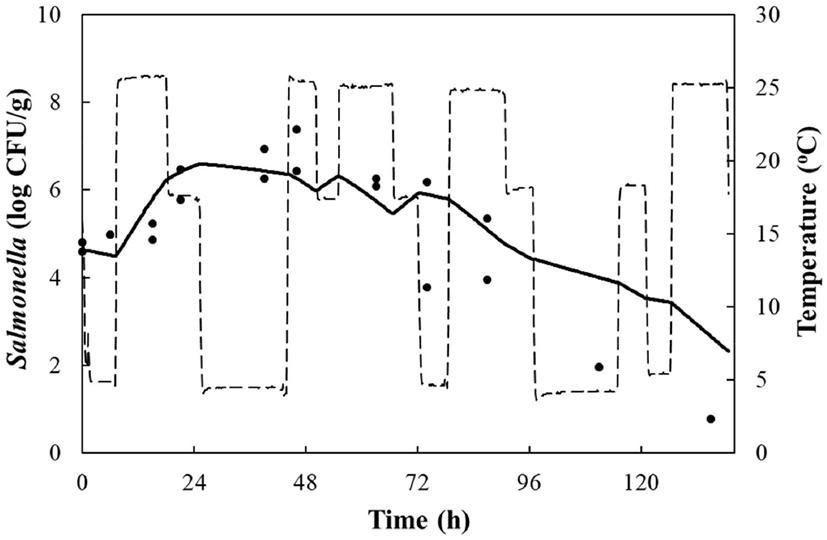
In conclusion, if Salmonella contaminates napa cabbage kimchi during manufacture, the pathogen can multiply for 17-24 h during fermentation, but perishes at 15-30°C due to an increase in LAB. However, Salmonella can resist or grow slowly at 4°C and 10°C. Therefore, napa cabbage kimchi should be consumed at least after 384 h of fermentation at 4°C, and at least after 48 h of fermentation at 25-30°C. In addition, the developed models can be applied to describe or predict the kinetic behavior of Salmonella in napa cabbage kimchi during fermentation.










