Article
제품화된 18종 동결건조 프로바이오틱스의 동정을 위한 RAPD-PCR 분석법 평가
이보솜
1, 양수연
2, 최보영
2, 이민지
2, 반오현
1,2, 양정우
2,*
, 정영훈
1,3,*
Evaluation of randomly amplified polymorphic DNA-polymerase chain reaction method for the identification of eighteen lyophilized probiotics commercially available
Bo Som Lee
1, Soo-Yeon Yang
2, Boyoung Choi
2, Minjee Lee
2, O-Hyun Ban
1,2, Jungwoo Yang
2,*
, Young Hoon Jung
1,3,*
1School of Food Science and Biotechnology, Kyungpook National University, Daegu 41566, Korea
2Ildong Bioscience, Pyeongtaek 17957, Korea
3Institute of Fermentation Biotechnology, Kyungpook National University, Daegu 41566, Korea
*Corresponding author. Jungwoo Yang. E-mail:
yjw@ildong.com, Phone:+82-31-646-3180, Fax:+82-31-070-2497 Young Hoon Jung. E-mail:
younghoonjung@knu.ac.kr, Phone:+82-53-953-9555, Fax:+82-53-950-5777
Copyright © The Korean Society of Food Preservation. This is an Open Access article distributed under the terms of the
Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0) which permits
unrestricted non-commercial use, distribution, and reproduction in any
medium, provided the original work is properly cited.
Received: Nov 11, 2020; Revised: Dec 21, 2020; Accepted: Apr 08, 2021
Published Online: Jun 30, 2021
요약
제품화를 위한 프로바이오틱스의 신속한 동정은 품질보증을 위한 중요한 단계이다. 이를 위해 다양한 동정 방법들이 시도되고 있으나, RAPD-PCR 분석법은 편리성 등의 장점으로 널리 오랫동안 사용되어 왔다. 본 연구에서는 18종 프로바이오틱스의 동정을 위하여 RAPD-PCR 분석법을 적용하였고, 이를 기반으로 제품형태를 블라인트 테스트한 결과, 18종 균주별로 정확히 동정됨을 확인할 수 있었다. 그리하여, 본 연구에서 적용된 RAPD-PCR 분석법은 제품화된 프로바이오틱스의 품질표준을 위한 적용이 가능함을 시사한다.
Abstract
Identification of probiotics is crucial to ensure the quality of food products manufactured at the industrial scale. Although various techniques have been introduced for bacterial identification, randomly amplified polymorphic DNA-polymerase chain reaction (RAPD-PCR) is generally accepted as a conventional method for bacterial identification. In this study, RAPD-PCR method was evaluated for the identification of eighteen commercial probiotic strains. In results, cellular and product (lyophilized) forms of each probiotic strain were successfully identified based on band intensities and size of the amplified genes. Therefore, RAPD-PCR method may be applied for the identification of lyophilized probiotics commercially available.
Keywords: probiotics; lactic acid bacteria; randomly amplified polymorphic DNA; RAPD
Introduction
Due to the recent escalating interest in health foods, considering their health benefits, such as immune-boosting, cholesterol-lowering, and anti-inflammatory effects (Granato et al., 2010), the probiotics market has gradually expanded (de Simone, 2019; Kechagia et al., 2013). Quality control of probiotics produced on a commercial scale is important to ensure that safe and reliable products are provided to the consumers. Typically, this includes identification of probiotic bacteria, and detection of contaminants and toxic compounds. Although various techniques have been introduced for bacterial identification (e.g., 16S rRNA sequencing, restriction fragment length polymorphism, rep-PCR, and ribotyping) (Mohania et al., 2008; Yu et al., 2009), randomly amplified polymorphic DNA-polymerase chain reaction (RAPD-PCR) analysis is regarded as the standard method (Booysen et al., 2002; Temmerman et al., 2004). The principle of RAPD-PCR is that oligonucleotide primers bind to different loci in the genome of a bacterium and randomly amplify unique fragments (Williams et al., 1990). This method is used to reveal the overall differences or similarities among interspecies or intraspecies (Jarocki et al., 2020; Sharma et al., 2020).
In this study, RAPD-PCR was applied for the rapid identification of eighteen commercial probiotic strains. First, the most suitable primer was screened among three universal primers (5'-ACGCAGGCA-3', 5'-ACGAGGCAC-3', and 5'- ACGCGCCCT-3'). Then, RAPD-PCR patterns of cellular and product (lyophilized) forms were compared to determine uniformity. Results from this study suggest that the present RAPD-PCR method may be applied for the identification of lyophilized probiotics commercially available.
Materials and methods
Bacterial strains and culture conditions
The bacterial strains used in this study are listed in Table 1. The bacterial cells were incubated in de Man, Rogosa, and Sharp (MRS) broth (BD Difco, Franklin Lakes, NJ, USA) at 37°C aerobically in a static incubator. Bacillus coagulans IDCC 1201 and Lactococcus lactis IDCC 2301 were incubated in a shaking incubator at 200 rpm and 45°C and 37°C, respectively.
Table 1.
Bacterial strains used in this study
|
Strain |
Origin |
ATCC accession no. |
|
Enterococcus faecium IDCC 2102 |
Infant feces |
BAA-3146 |
|
Streptococcus thermophilus IDCC 2201 |
Homemade yogurt |
BAA-3150 |
|
Lactococcus lactis IDCC 2301 |
Homemade cheese |
BAA-2834 |
|
Lactobacillus gasseri IDCC 3101 |
Breast milk |
BAA-2841 |
|
Lactobacillus rhamnosus IDCC 3201 |
Infant feces |
BAA-2836 |
|
Lactobacillus acidophilus IDCC 3302 |
Infant feces |
BAA-2845 |
|
Lactobacillus casei IDCC 3451 |
Infant feces |
BAA-2843 |
|
Lactobacillus plantarum IDCC 3501 |
Kimchi (fermented food) |
BAA-2838 |
|
Lactobacillus salivarius IDCC 3551 |
Healthy child saliva |
BAA-2835 |
|
Lactobacillus reuteri IDCC 3701 |
Breast milk |
BAA-2837 |
|
Lactobacillus helveticus IDCC 3801 |
Infant feces |
BAA-2840 |
|
Lactobacillus fermentum IDCC 3901 |
Homemade cheese |
BAA-2842 |
|
Bifidobacterium longum IDCC 4101 |
Infant feces |
BAA-2847 |
|
Bifidobacterium bifidum IDCC 4201 |
Infant feces |
BAA-2850 |
|
Bifidobacterium lactis IDCC 4301 |
Infant feces |
BAA-2848 |
|
Bifidobacterium breve IDCC 4401 |
Infant feces |
BAA-2849 |
|
Lactobacillus johnsonii IDCC 9203 |
Infant feces |
BAA-3147 |
|
Bacillus coagulans IDCC 1201 |
Green malt |
BAA-3143 |
Download Excel Table
Randomly amplification polymorphic DNA
Genomic DNA from cellular and lyophilized form of probiotics was isolated using the WizardTM genomic DNA purification kit (Promega, Madison, USA). Briefly, lyophilized probiotics were washed twice with ddH2O to remove excipients prior to DNA isolation. PCR was performed with the universal primers: 5'-ACGCAGGCA-3' (Sesena et al., 2004); 5'-ACGAGGCAC-3' (Tilsala-Timisjarvi and Alatossava, 1998); and 5'-ACGCGCCCT-3' (Johansson et al., 1995). Randomly amplified PCR products were analyzed and visualized using a GelDoc imaging system (Bio-Rad, Hercules, CA). Finally, the products were quantified using Image Lab software (Bio-Rad, version 6.1) for further analysis.
Results and discussion
Selection of a suitable primer for RAPD-PCR analysis
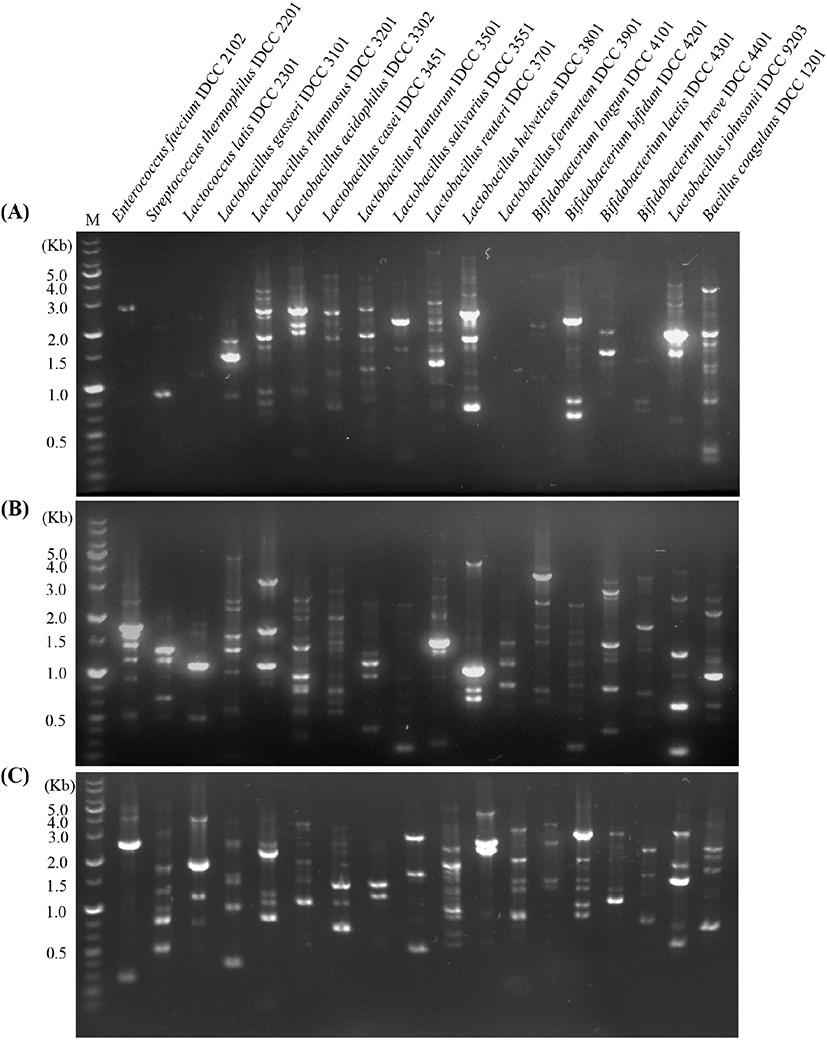
Prior to identification using the RAPD-PCR analysis, identification of eighteen probiotic strains was verified by 16S rRNA sequence analysis (data not shown). Next, three universal primers were evaluated and the most suitable primer was selected. Unique and distinctive band patterns of the eighteen species were clearly observed based on band intensities and gene size (Fig. 1). However, when performing PCR using P1 primer, faint bands were observed for Enterococcus faecium IDCC 2102, Streptococcus thermophilus IDCC 2201, L. lactis IDCC 2301, Bifidobacterium longum IDCC 4101, B. breve IDCC 4401, and Lactobacillus fermentum IDCC 3901 (Fig. 1A). When performing the analysis using P2 primer, it was difficult to distinguish L. salivarius IDCC 3551 from B. bifidum IDCC 4201 due to the absence of a major band. Finally, P3 primer was selected as the suitable primer as it showed significant differences between the eighteen probiotic strains tested in the present study (Fig. 1C). These results are consistent with previous reports which showed differences or similarities among interspecies through RAPD-PCR analysis (Jarocki et al., 2020; Sharma et al., 2020).
Fig. 1.
Evaluation of three universal primers for RAPD-PCR analysis.
A, P1 (5'-ACGCAGGCA-3'); B, P2 (5'-ACGAGGCAC-3'); C, P3 (5'-ACGCGCCCT-3') primers. The figure is the representative from three independent experiments.
Download Original Figure
Rapid identification of eighteen lyophilized probiotics commercially available
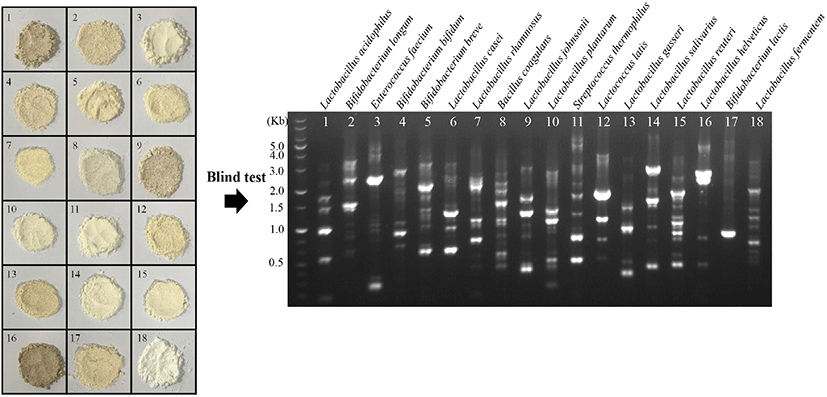
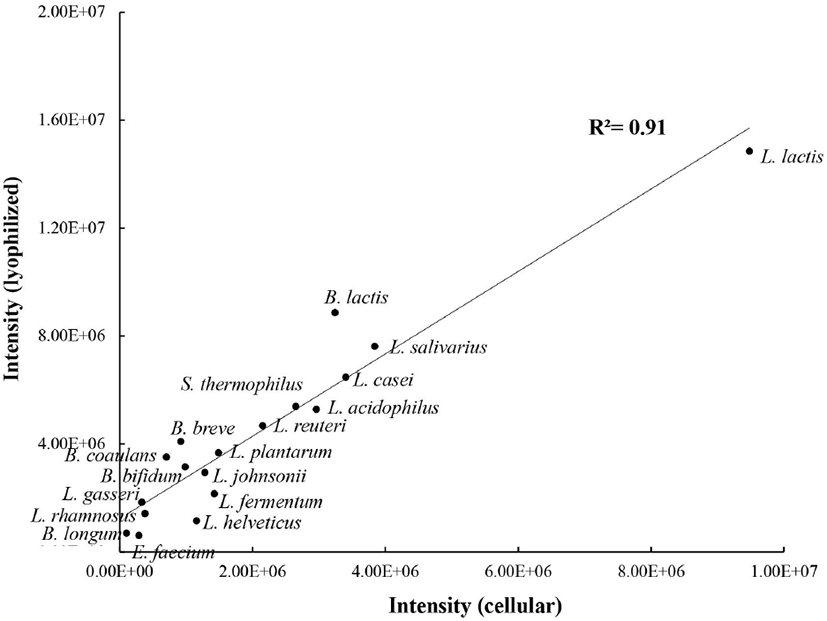
Based on the RAPD-PCR patterns using primer P3, a blind test for the rapid identification of lyophilized eighteen probiotics was performed. The RAPD-PCR profiles of the lyophilized form of probiotics were consistent with those of the cellular form (Fig. 1C and 2). For example, the two bands (∼2.5 kb and ∼0.3 kb) in lane 3 of sample No. 3 were identified as E. faecium IDCC 2102. One major band (∼1.8 kb) and three minor bands (∼4.0 kb, ∼1.2 kb, and ∼0.8 kb) in lane 12 were identified as L. lactis IDCC 2301. Two major bands (∼3.0 kb and ∼2.7 kb) and one minor band (∼5.0 kb) in lane 16 were identified as L. helveticus IDCC 3801. Finally, the correlation between cellular and lyophilized forms of probiotics was determined bassed on the intensities of the major bands observed only for a specific strain (Fig. 1C and 2) using Image Lab software (Bio-Rad, version 6.1). The coefficient of determination (R2) was 0.91, indicating the RAPD-PCR method used in this study was reliable for the identification of the eighteen lyophilized probiotic strains commercially available (Fig. 3).
Fig. 2.
Blind test for identification of eighteen lyophilized probiotics commercially available using RAPD-PCR analysis.
The figure is the representative from three independent experiments.
Download Original Figure
Fig. 3.
orrelation of RAPD-PCR patterns between cellular and lyophilized forms of probiotics.
A, P1 (5'-ACGCAGGCA-3'); B, P2 (5'-ACGAGGCAC-3'); C, P3 (5'-ACGCGCCCT-3') primers. The figure is the representative from three independent experiments.
Download Original Figure
Acknowledgments
This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through High Value-added Food Technology Development Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (321033-3) and a National Research Foundation of Korea (NRF) funded by the Korea government (MSIT) (No. 2020R1C1C1005251). This work was performed at the Ildong Bioscience Research Center and School of Food Science and Biotechnology in Kyungpook National University.
References
Booysen C, Dicks LMT, Meijering I, Ackermann A. Isolation, identification and changes in the composition of lactic acid bacteria during the malting of two different barley cultivars. Int J Food Microbiol, 76, 63-73 (2002)


Jarocki P, Komon-Janczara E, Glibowska A, Dworniczak M, Pytka M, Korzeniowska-Kowal A, Wzorek A, Kordowska- Wiater M. Molecular routes to specific identification of the
Lactobacillus casei group at the species, subspecies and strain level. Int J Mol Sci, 21, 2694 (2020)




Johansson ML, Quednau M, Molin G, Ahrne S. Randomly amplified polymorphic DNA (RAPD) for rapid typing of
Lactobacillus plantarum strains. Lett Appl Microbiol, 21, 155-159 (1995)



Mohania D, Nagpal R, Kumar M, Bhardwaj A, Yadav M, Jain S, Marotta F, Singh V, Parkash O, Yadav H. Molecular approaches for identification and characterization of lactic acid bacteria. J Dig Dis, 9, 190-198 (2008)



Schillinger U, Yousif NMK, Sesar L, Franz CMAP. Use of group-specific and RAPD-PCR analyses for rapid differentiation of
Lactobacillus strains from probiotic yogurts. Curr Microbiol, 47, 453-456 (2003)



Temmerman R, Huys G, Swings J. Identification of lactic acid bacteria: Culture-dependent and culture-independent methods. Trends Food Sci Technol, 15, 348-359 (2004)