1. Introduction
The Centers for Disease Control (CDC) reported that Escherichia coli, Salmonella, and Shigella were considered major foodborne pathogens that caused 50,390 outbreaks with 8,010 hospitalizations during 2009-2020 in the USA (CDC, 2022). These foodborne pathogens are commonly prevalent in foods such as ground beef, dairy products, chicken, and eggs (Duc et al., 2020; Rangel et al., 2005; Shahin and Bouzari, 2018), and recently in fresh produce (Bai et al., 2019; Ruengvisesh et al., 2019). Several physical and chemical methods have been introduced and utilized in fresh produce to control foodborne pathogens using a thermal treatment (Schweiggert et al., 2007), irradiation (Molins et al., 2001), chlorine treatment (Petri et al., 2015), and ozone treatment (Chaidez et al., 2007). The thermal treatment, popularly used in meat and meat products, is limited when employed in fresh produce due to the loss of nutrients and freshness (Fan et al., 2017). In addition, irradiation also led to color and texture changes, lipid oxidation, unpleasant odor, and poor consumer acceptability (Duc et al., 2020). Likewise, treatments with chlorine and ozone have been reported due to the possibility of the development of hazardous and carcinogenic compounds such as trihalomethanes and chlorite in the produce and detrimental health effects upon exposure to high concentrations of ozone (Bai et al., 2019). Thus, the negative impacts of these physical and chemical methods have increased the need to search for and develop a novel biocontrol agent that is safe and eco-friendly without affecting the nutrients and quality of fresh produce.
The exploration of phages as biocontrol agents has increased as a natural and green technology for effectively controlling of foodborne pathogens in fresh produce (Oh and Park, 2017). Phage is a virus that can infect and kill target bacteria specifically (Zampara et al., 2017). A phage exhibits either lytic or lysogenic characteristics depending on the life cycle of a phage with host bacteria (Clokie et al., 2011; Gill and Hyman, 2010). The lytic phage rapidly replicates within the infected bacterial chromosome and eventually lyses the cells, resulting in the host’s death. In contrast, the lysogenic phage can exist as a prophage within the host cell after its DNA integration into the bacterial chromosome and does not result in the lysis of the bacterial cells. Thus, lytic phage has been the preferred control of foodborne pathogens due to the ability to specifically infect and eventually lyse the host bacteria (Bai et al., 2019). Recently, several lytic phages have been investigated as biocontrol agents; however, most of them could infect only one bacterial genus (Moye et al., 2018; Oh and Park, 2017; Wei et al., 2019). The narrow host range of phage becomes limited for the control of several foodborne pathogens simultaneously in fresh produce. Hence, development of phages with broad host range is more desirable and effective as biocontrol agents since they can simultaneously infect multiple bacterial genera (Ross et al., 2016). Although phages specific to either Salmonella, Shigella, or E. coli O157:H7 have been reported in both in vitro and in vivo experiments (Oh and Park, 2017; Sabouri et al., 2017; Wei et al., 2019), a lytic phage that could simultaneously infect multiple foodborne pathogens is rarely reported (Table S1). This study investigated that a lytic phage KFS-EC3 efficiently infects E. coli O157:H7, Shigella sonnei, and Salmonella Mission. Our findings lay the groundwork for providing further insights into phage studies for the simultaneous control of foodborne pathogens contaminating a food product.
The safety, lytic property, and novelty of the phage should be investigated by genomic analysis to be able to use phages as biocontrol agents (Newase et al., 2019; Sritha and Bhat, 2018). Whole genome sequencing provides information on the genetics of the isolated phages and highlights the safety of the phage usage by confirming the absence of genes encoding lysogenic, virulence, antibiotic, and allergenic factors in the genome. Furthermore, genomic characteristics can be compared to those of other reported phages to ascertain the novelty of the phage. Therefore, this study aimed to characterize the broad host range and bactericidal effect, and to genetically analyze the safety, novelty, and lytic property of the phage KFS-EC3 as a biocontrol agent.
2. Materials and methods
Escherichia coli O157:H7 ATCC 10536 was obtained from the American Type of Culture Collection (ATCC) and used as an indicator strain for phage propagation. E. coli O157:H7 ATCC 10536 was cultivated in 25 mL of tryptic soy broth (TSB, Difco Laboratories Inc., Sparks, MD, USA) at 37°C for 16 h with gentle shaking. After washing three times with sterilized phosphate-buffered saline (PBS 1×, pH 7.4, Life Technologies Co., Carlsbad, CA, USA) by centrifuging at 7,000 ×g for 4 min at 4°C, the bacterial pellet was resuspended with PBS. The concentration of the bacterial suspension was adjusted to 8 log CFU/mL based on standard curves previously constructed by measuring optical density at 640 nm.
Phage KFS-EC3 was previously isolated from a slaughterhouse sewage sample (Daegu, Korea) (Kim et al., 2021). For its high-titer propagation, 1% (v/v) of E. coli O157:H7 ATCC 10536 suspension was mixed with 3 mL of TA broth (8 g/L nutrient broth, 5 g/L NaCl, 0.2 g/L MgSO4, 0.05 g/L MnSO4, and 0.15 g/L CaCl2), and the mixture was incubated at 37°C for 2 h. Subsequently, 1 mL of phage solution was added followed by incubation for 2 h at 37°C. After centrifuging the mixture at 6,000 ×g for 20 min at 4°C, the supernatant was filtered using a 0.20-μm cellulose acetate filter. This procedure was repeated with increasing amounts of TA broth. The final concentration of the propagated phage was measured using the plaque assay as follows: TA soft agar (0.4% agar) including 100 μL of the phage and 200 μL of the bacterial suspension (∼8 log CFU/mL) was poured onto TSA plates and incubated at 37°C for 16 h. Afterward, phage purification was performed using the previously described method (Lee et al., 2020). The propagated phage was mixed with 10% (w/v) polyethylene glycol 6000 (Sigma-Aldrich Co., St. Louis, MO, USA) and 10 mL of 1 M NaCl, and then precipitated by incubating at 4°C for 16 h. After centrifugation at 7,200 ×g for 20 min at 4°C, the obtained phage pellet was suspended in sodium chloride-magnesium sulfate (SM) buffer (50 mM/L Tris-HCl, 100 mM NaCl, and 10 mM MgSO4, pH 7.5). CsCl-gradient ultracentrifugation was then performed at 39,000 ×g for 2 h at 4°C. A bluish opalescent band was collected, dialyzed, and stored in a glass vial at 4°C for further characterization studies. The final concentration of the purified phage (hereafter referred to as KFS-EC3) was determined using the plaque assay expressed as a plaque-forming unit (PFU)/mL. After phage purification, plaque assay was performed against host strains of E. coli ATCC 10536, Shigella sonnei ATCC 9290, and Salmonella Mission to confirm the host specificity of the phage.
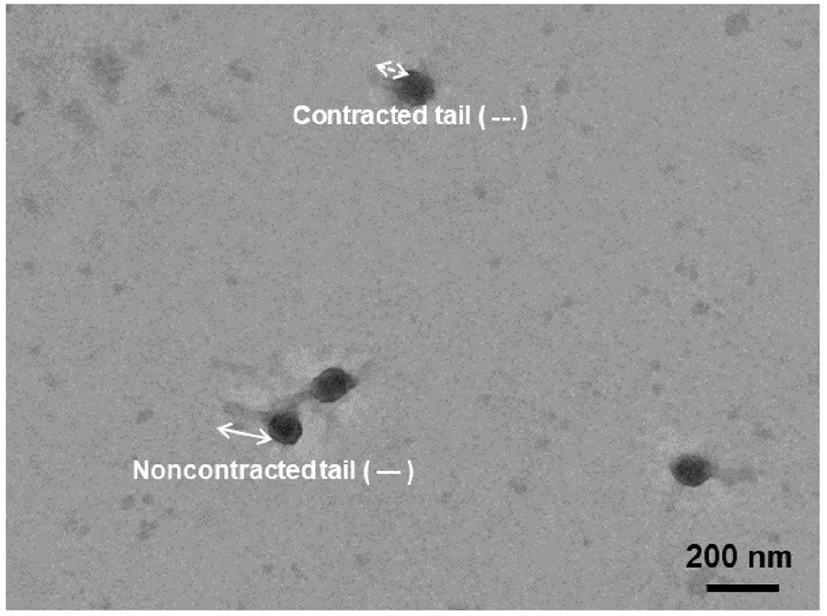
The morphological characteristics of KFS-EC3 were previously observed (Kim et al., 2021), however, a contractile property of its tail was found in this study. For transmission electron microscopy (TEM, H-7100, Hitachi Ltd., Tokyo, Japan) analysis, a carbon-coated copper grid was dipped into 1 mL of KFS-EC3 for 1 min and then stained with 2% uranyl acetate (Sigma-Aldrich Co.). The phage on the grid was observed at an accelerating voltage of 100 kV with 40,000× magnification.
The phage was added to each bacterial suspension (∼8 log CFU/mL) of E. coli O157:H7 ATCC 10536, S. Mission, S. sonnei ATTC 9290, and their cocktail (1:1:1, v/v) at MOIs of 0.001, 0.1, and 1.0 to evaluate the polyvalent bactericidal activity of KFS-EC3. SM buffer was used for comparison in the control. The mixtures of the phage and bacterial suspension were incubated at 37°C for 4 h with gentle shaking. The viable bacterial concentration was determined by plate count method using xylose lysine deoxycholate (XLD, Difco Laboratories Inc.) and eosin methylene blue (EMB, Difco Laboratories Inc.) agar plates.
The genomic DNA of KFS-EC3 was extracted using a Phage DNA Isolation Kit (Norgen Biotek Corp., Thorold, ON, Canada) according to the manufacturer’s instructions. The genome sequencing was performed using the Illumina Miseq platform with paired-end reads of 2×150 bp size. The sequenced reads were assembled to single contig using the SPAdes genome assembler (Illumina Inc., San Diego, CA, USA). The complete sequence of KFS-EC3 genome was available in the GenBank database under accession number MZ06535. The contig of KFS-SE3 genome was annotated using the Rapid Annotations Systems Technology (RAST) server (Aziz et al., 2008). The genome was analyzed for genes involved in host lysis and prophage to confirm the lytic cycle of KFS-EC3, using BLASTp, conserved domain database, and PHASTER (Arndt et al., 2016). In addition, potential virulence factors and antibiotic resistance genes were screened against the virulence factor database (Liu et al., 2022) and Comprehensive Antibiotic Resistance Database (https://card.mcmaster.ca, 5 November 2022), respectively.
The phylogenetic analysis was performed to confirm the classification and novelty of KFS-EC3, followed by a comparison of genomic characteristics with phylogenetically related phages. The amino acid sequences of similar myophages’ tail proteins were obtained from the NCBI database and aligned using the ClustalW algorithm (Larkin et al., 2007). The UPGMA tree was constructed based on tail protein using Splits Tree4 (Huson and Bryant, 2010) with 1,000 bootstrap replicates. Additionally, the general genomic characteristics, identity, and ANI value of phylogenetically related phages were confirmed using the NCBI database, BLASTN analysis, and OrthoANI Tool v0.93 (Lee et al., 2016b), respectively.
Experiments were conducted in triplicate and the data were expressed as mean±standard deviation (SD). Statistical analysis was performed using InStat V.9 (GraphPad, San Diego, CA, USA). Student’s paired t-test and one-way analysis of variance (ANOVA) were used to compare in-between groups and among groups, respectively. Differences among means were analyzed using Ducan’s Multiple Range Test (DMRT), and the significance level was defined at p⟨0.05.
3. Results and discussion
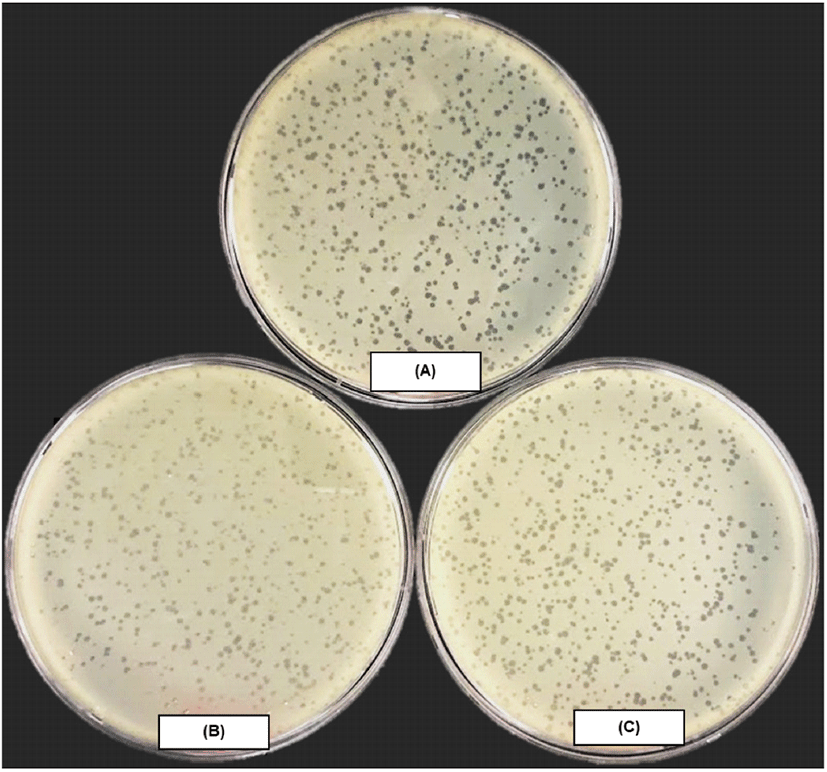
KFS-EC3, a previously isolated phage, was propagated and purified at a concentration of (2.3±0.2)×1011 PFU/mL. As shown in Fig. 1, small and clear plaques of KFS-EC3 were observed on the lawn of E. coli ATCC 10536, S. sonnei ATCC 9290, and S. Mission, indicating its specificity with broad host range. Although there is no clear definition of between broad and narrow host range of phage, broad host range was often referred to phages infecting more than two bacterial genera (Bielke et al., 2007; Kim et al., 2021). The broad host range of KFS-EC3 had benefits for practical use to control multiple foodborne pathogens in food, rather than reported phages with a narrow host range such as phage Phaxl (Shahrbabak et al., 2013), phage phiLLS (Amarillas et al., 2017), and phage vB_EcoS_HSE2 (Peng and Yuan, 2018). Similar to this study, several studies have also showed lytic E. coli phages with broad host range (Table S1). Most of them can infect two bacterial genera such as phages PS5 (Duc et al., 2020), SFP10 (Park et al., 2012), vB_EcoM_swi3 (Sui et al., 2021), phiC119 (Amarillas et al., 2016), HY01 (Lee et al., 2016a) and SH6 (Hamdi et al., 2017), which means a narrower host range than the phage KFS-EC3. Some E. coli phages capable of infecting three genera have also been reported, such as phage EcS1 (Saad et al., 2019) and SH7 (Hamdi et al., 2017) specific to E. coli, Salmonella spp., and Shigella spp. Overall, a broad host range at the genus level of KFS-EC3 would be advantageous for the simultaneous biocontrol of E. coli O157:H7, Salmonella, and S. sonnei in food.
The morphological analysis by TEM showed that the KFS-EC3 phage had an icosahedral head with a length of 97.1±5.2 nm and a contractile tail with a length ranging from 67.2 to 135.5 nm (Fig. 2). KFS-EC3 was found to be myophage based on these characteristics. Compared with other reported myophages with broad host ranges (Table S1), the head and tail lengths of KFS-EC3 phage were longer than those of phage PS5 (Duc et al., 2020) and vB_EcoM_swi3 (Sui et al., 2021). Additionally, KFS-EC3 had a shorter head and longer tail length than that of phage HY01 (Lee et al., 2016a), EcS1 (Saad et al., 2019), and SH7 (Hamdi et al., 2017). Thus, our findings suggested that KFS-EC3 exhibits broad host range and unique morphological differences from other myophages reported infecting E. coli.
The polyvalent bactericidal activity of KFS-EC was evaluated and compared against each single host strain and their bacterial cocktail at various MOIs (Fig. 3). After phage treatment for 4 h at all MOIs, significant reductions of each single host strains and their bacterial cocktail were observed compared to the control (p⟨0.05). Bactericidal activities of KFS-EC3 at the lowest MOI of 0.001 were more outstanding than at other MOIs of 0.1 and 1.0 with bacterial reductions of 4.1±0.1, 3.4±0.2, and 3.1±0.2 log CFU/mL against E. coli O157:H7, S. sonnei, and S. Mission, respectively. In addition, the bacterial reduction of E. coli O157:H7 was significantly greater than those of S. sonnei, and S. Mission at MOIs of 0.001 and 0.1. However, there are no significant differences in bacterial reduction of them at MOI of 1.0. Interestingly, KFS-EC3 also exhibited competitive reductions of bacterial cocktail by ∼3.5± 0.3 log CFU/mL reduction at all MOIs. Thus, KFS-EC3 showed excellent polyvalent bactericidal activity when exposed to both single-host bacteria and multi-host bacteria.
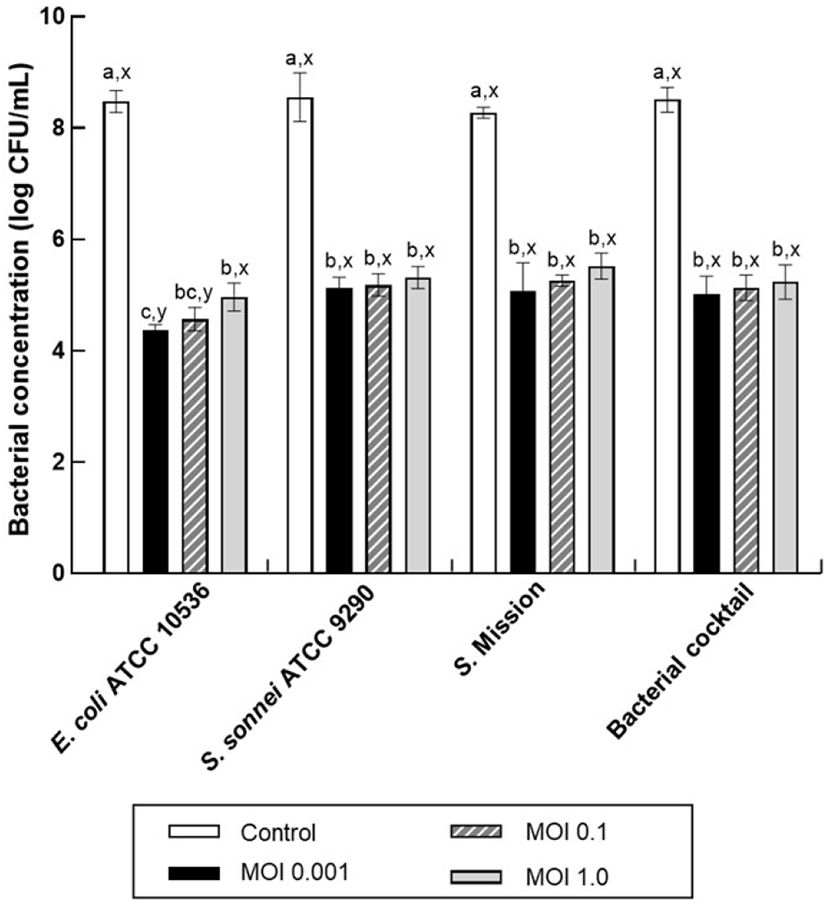
Similarly, a previous study (Lee et al., 2016a) reported that phage HY01 reduced each host bacteria of E. coli O157:H7 ATCC 43901 and S. flexneri 2a strain 2457T by ∼4 log CFU/mL at MOI of 10. However, since fresh produce can be contaminated with several pathogens (Alegbeleye et al., 2018), the polyvalent bactericidal activity of the phage against bacterial cocktail was critical for simultaneous biocontrol of the foodborne pathogens in food application. Park et al. (2012) reported that the polyvalent bactericidal activity of phage SFP10 against a bacterial cocktail of S. Typhimurium and E. coli O157:H7, which simultaneously reduced S. Typhimurium by 1.8 log CFU/mL and E. coli by 4.3 log CFU/mL at MOI of 100. Duc et al. (2020) also showed that phage PS5 effectively reduces S. Enteritidis, S. Typhimurium, E. coli O157:H7, and their bacterial cocktail. Phage PS5 treatment at MOI of 10,000 caused significant bacterial reduction by ∼2 log CFU/mL for S. Enteritidis, S. Typhimurium, and bacterial cocktail and by ∼4 log CFU/mL for E. coli O157:H7. Compared to those of other E. coli phages with broad host specificity, KFS-EC3 exhibited similar polyvalent bactericidal activity at lower MOI. A lower MOI of the phage has been reported to be more beneficial and economical for use in food application (Oh and Park, 2017). Our study revealed that KFS-EC showed excellent polyvalent bacterial inhibition against multiple foodborne pathogens and their cocktail even at a very low MOI of 0.001. These results indicate the potential of KFS-EC3 as a biocontrol agent with great effectiveness and efficiency for controlling foodborne pathogens.
The genome of KFS-EC3 consisted of 166,440 bp with a G+C content of 35.5%. Its genome comprised 122 hypothetical ORFs, 151 functional ORFs, and 8 tRNAs (Table 1). The annotated functional genes were categorized into five functional groups: including nucleotide metabolism, phage packaging, structure, host lysis, and additional functions (Table S2). Fifty-nine genes were confirmed to participate in nucleotide metabolism, such as RNA polymerase, DNA polymerase, DNA ligase, DNA helicase, ribonucleotide reductase, exo/endonuclease, transcriptional regulator, etc. They have a role in DNA replication, recombination, repair, and their regulation. Furthermore, sixteen genes involved in phage packaging were found, such as porter protein (ORF 115), phage prohead assembly and capsid scaffolding proteins (ORF 38, 42, 91, 107, 111-114, and 133), phage tail assembly and scaffolding proteins (ORF 95, 120, 136, and 138), and terminase (ORF 118 and 119). Especially, terminase is known to play an essential role in the initiation of phage packaging by DNA recognition, cleavage, and its translocation into phage prohead (Rao and Feiss, 2008; Shen et al., 2012).
The main components of the phage structure were also found at the posterior of the genome and predicted to encode capsid (ORF 86, 109-110, and 139), neck (ORF 121 and 122), tail (ORF 69-73, 89-90, 116-117, and 123-124), and baseplate (ORF 96-102, 125-129, 131, and 132). Among these components of phage structure, tail fibers on the side of the phage tail can lead to phage attachment to the host bacterial membrane and its DNA injection, which are responsible for determining phage host specificity (Garcia-Doval and van Raaij, 2012). Interestingly, tail fiber proteins from ORF 70, 71, and 124 of KFS-EC3 were similar to those of E. coli phages PaulHMueller (QXV83448.1, 76.9%), ime09 (YP_007004622.1, 100.0%), and T6 (YP_010067303.1, 100.0%), respectively. Other fiber proteins from ORF 69, 72, and 73 were similar to those of Shigella phages A2 (UVD36934.1, 98.9%), KRT47 (QHB43271.1, 98.7%), and Esh26 (URY13523.1, 98.1%), whereas proteins from ORF 89 and 90 were similar to those of Salmonella phages Lv5cm (QVW09019.1, 98.7%) and SG1 (YP_010075104.1, 98.0%). These results agreed with the conserved domain analysis of phage tail fibers (Table 2). Finally, our results suggested that these multiple tail fibers of KFS-EC3 may contribute to its broad host range against E. coli, Shigella, and Salmonella.
The lytic activity of KFS-EC3 was further evaluated by confirming host lysis proteins and prophage for use as a biocontrol agent. From the annotation and conserved domain analysis (Table 2 and S2), the genes involved in host lysis were identified as a holin (ORF 68) and endolysin (ORF 143). Phage endolysin can lyse the bacterial cell wall during the end of a lytic cycle for its release, which was supported by the holin to form holes in the bacterial membrane (Li et al., 2021). Most endolysins, which lyse gram-negative host bacteria, are structured to have a single domain of enzymatically active domain (EAD) (Abdelrahman et al., 2021). From the in silico analysis, the endolysin (ORF143) of KFS-EC3 belongs to the muramidase family comprised of one EAD, such as reported E. coli phages of T7 (Inouye et al., 1973), T4 (Matthews and Remington, 1974), and vB_EcoM_ECP26 (Park and Park, 2020). In addition, KFS-EC3 did not contain any genes associated with prophage, virulent factors, antibiotic resistance, and allergenicity. Overall, KFS-EC3 exhibited its potential as a lytic and safe biocontrol agent for food application.
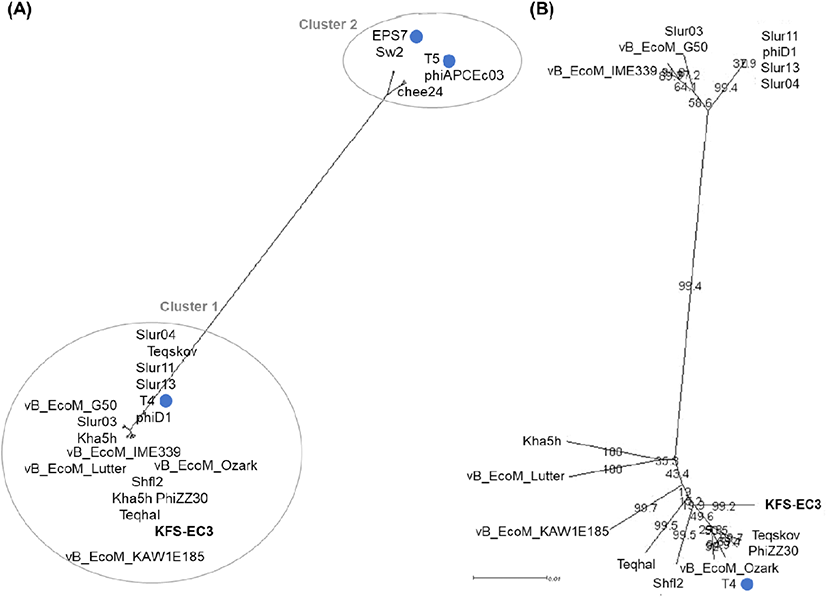
Phylogenetic analysis based on tail protein (Fig. 4(A)) revealed that similar E. coli myophages were divided into two groups. KFS-EC3 was clustered with 16 phages from the genus Tequatrovirus of the family Myoviridae (indicated as cluster 1), whereas cluster 2 contained phages of other genera Tequintavirus (phages T5, chee24, and phiAPCEc03) and Epseptimavirus (phages EPS7 and Sw2). In further comparison between phages of cluster 1 (Fig. 4(B)), KFS-EC3 was particularly related to 9 phages, including type phage T4 of E. coli phage. Then, genomic features, identity, and ANI value of KFS-EC3 were compared with these phylogenetically similar 9 phages (Table 1). These phages have a similar genome size of ∼165-170 kbp, GC contents of ∼35-40%, and tRNA of 6-10. A total of tRNAs in the KFS-EC3 genome was greater than phage Teqskov and similar to phages vB_EcoM_Lutter and T4. The presence of tRNA genes in the phage genome has been reported to enhance the protein translation efficiency in host bacteria and/or expand the host specificity (Grose et al., 2014; Morgado and Vicente, 2019). ANI analysis showed that KFS-EC also have a genetically close relationship with phages Teqskov (95.73%), Shfl2 (95.64%), and vB_EcoM_Ozark (95.47%). These ANI values were higher than the mechanistic demarcation criterion (70%) for phage genera (Turner et al., 2021). Overall, KFS-EC3 was classified as a new member of the Tequatrovirus genus of the Myoviridae family with genetically different characteristics from other reported phages.
4. Conclusions
Phage-based biocontrol strategy is a promising approach for controlling foodborne pathogens in fresh produce. This study investigated the suitability of phage KFS-EC3 on the control of multiple foodborne pathogens and its genomic characteristics for use as a biocontrol agent. KFS-EC3 showed efficient and effective bactericidal effect to reduce E. coli O157:H7, S. sonnei, and S. Mission, as well as their bacterial cocktail, indicating its potential for simultaneous biocontrol of these foodborne pathogens in fresh produce. The lytic property of the phage was also identified via genetic in silico analysis, which identified its host cell lysis genes and revealed the lack of genes related to prophage. Genomic in silico analysis revealed the safety of KFS-EC3 based on the absence of any genes associated with prophages, virulent factors, antibiotic resistance, and allergenicity. Furthermore, morphology and phylogenetic analyses revealed that KFS-EC3 is a new phage belonging to the Myoviridae family, Tequatrovirus genus. Therefore, the lytic and safe phage KFS-EC3 is a new, promising candidate of biocontrol agent for use in fresh produce. Future studies will evaluate the control efficacy of the phage against these foodborne pathogens in fresh produce. It is also important to evaluate its stability on the surface of fresh produce, which would highlight its potential for application to the washing and sanitizing process of fresh produce.










